I make an energy decomposition analysis in the complex LaCl3H2O. The structure has been optimized. By performing the SAPT calulation, I want to evaluate intermolecular interaction energy between LaCl3 and H2O. The input file is shown below,
memory 200000 MB
molecule dimer {
0 1
La -1.245901 -0.030094 0.049950
Cl -2.495563 2.194252 -0.517413
Cl 1.386678 -0.062614 0.280512
Cl -2.525909 -2.306411 -0.142389
0 1
O -0.908811 -0.341351 2.578561
H 0.039773 -0.403213 2.763659
H -1.365765 -0.963412 3.157789
}
dimerdist= 2.569900
set {
basis def2-qzvpp-ri
scf_type DF
freeze_core True
}
energy(‘sapt2+(3)dmp2’)
E_disp = get_variable(‘SAPT DISP ENERGY’) * psi_hartree2kcalmol
E_elst = get_variable(‘SAPT ELST ENERGY’) * psi_hartree2kcalmol
E_exch = get_variable(‘SAPT EXCH ENERGY’) * psi_hartree2kcalmol
E_ind = get_variable(‘SAPT IND ENERGY’) * psi_hartree2kcalmol
E_tot = get_variable(‘SAPT TOTAL ENERGY’) * psi_hartree2kcalmol
psi4.print_out("\n")
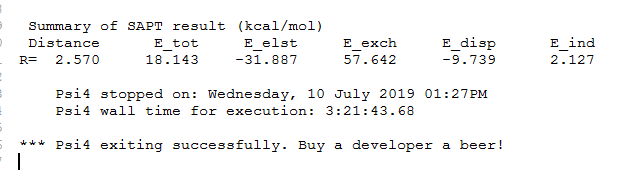
psi4.print_out(" Summary of SAPT result (kcal/mol)\n")
psi4.print_out(" Distance E_tot E_elst E_exch E_disp E_ind\n")
psi4.print_out("%s %6.3f %10.3f %10.3f %10.3f %10.3f %10.3f\n" % (“R=”,dimerdist,E_tot,E_elst,E_exch,E_disp,E_ind))
However, after calculation, it is found that the total binding energy is positive, as shown in the picture attachment. Is it because of the basis set used for La?
I want to know what is the reason and appreciate it If anyone knows how to solve it. Thanks!