Hi there.
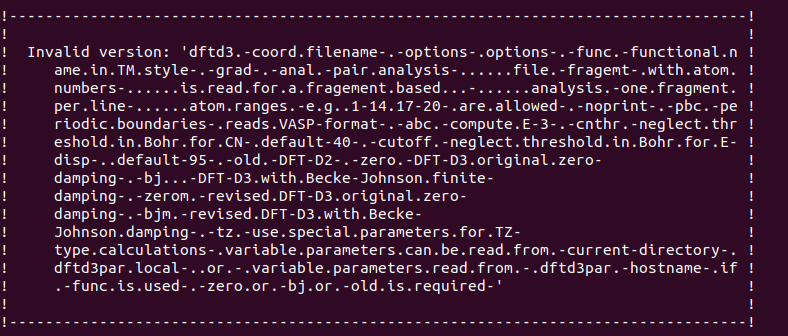
I’m using SAPT-0 and F/I-SAPT-0 to calculate the interaction energies between ligand-protein residue pairs. I am using the jun-cc-pvdz basis set with the d3 and d3mbj corrections to run the calculations, as well as scf_type df and freeze_core true. However, I am getting the print error message for both d3 and d3mbj calculations. I have already updated the Psi4 version to v. 1.8.0 and I am still getting this “Invalid Version” error, and I don’t know how to solve it anymore. Has anyone had a similar problem, and if so, how did you solve it?
I just updated Psi4 to 1.8.0 version, but this error still happening and I’m not sure about how to solve it.