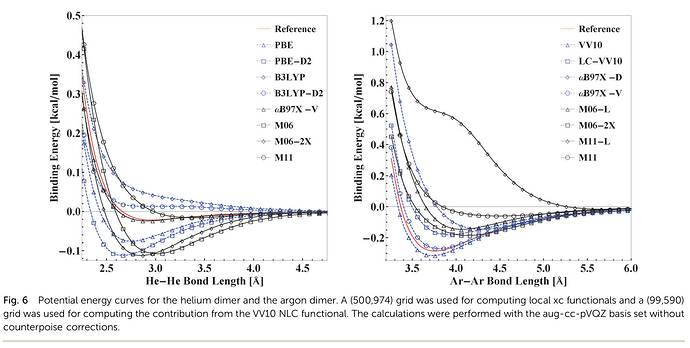
Dear all,
I’m trying to set-up the aforementioned functionals to compare the results to CP2K. I try to follow this publication for settings of the functionals in Psi4:
DOI: 10.1021/acs.jpca.8b03345
( Density Functional Theory for Microwave Spectroscopy of Noncovalent Complexes: A Benchmark Study)
to reproduce Ar-Ar interaction energies of the original wB97X-V paper (and possibly other energies):
DOI: 10.1063/1.4952647
(ω B97M-V: A combinatorially optimized, range-separated hybrid, meta-GGA density functional with VV10 nonlocal correlation)
here is the Python script that I use:
import numpy as np
import psi4
psi4.set_num_threads(8)
templ = """
0 1
Ar 0.0 0.0 0.0
Ar XXXX 0.0 0.0
nocom
noreorient
"""
Ar1 = """
0 1
Ar 0.0 0.0 0.0
nocom
noreorient
"""
method = "WB97M-V/def2-TZVPD"
method = "WB97X-V/def2-QZVPD"
psi4.set_options({"basis_guess": 0, "full_hess_every": hessian})
psi4.set_options({'scf_type':'df',"cc_type": "df"})
psi4.set_options({"dft_spherical_points": 1454, "dft_radial_points": 500})
psi4.set_options({"damping_percentage": 40})
psi4.set_options({"dft_bs_radius_alpha": 1.3})
psi4.set_options({"g_convergence": "interfrag_tight"})
energies = []
for x in range(0, 31):
cx = 3.25 + x*0.1
Ar2 = templ.replace("XXXX", str(cx))
mol = psi4.geometry(Ar2)
e2,wfn = psi4.energy(method, molecule=mol, return_wfn=True)
energies.append(e2 * psi4.constants.hartree2kcalmol)
energies = np.array(energies)
mol = psi4.geometry(Ar1)
e1,wfn = psi4.energy(method, molecule=mol, return_wfn=True)
e1 = e1 * psi4.constants.hartree2kcalmol
energies - 2*e1
the results are:
[0.26051278, -0.04387099, -0.22114193, -0.33644522, -0.39150897,
-0.40188769, -0.40330375, -0.00466373, -0.36437939, -0.34776137,
-0.30978748, -0.28900184, -0.25777778, -0.2334124 , -0.2097273 ,
-0.18432839, -0.17304105, -0.15599188, -0.14514117, -0.12967334,
-0.1173416 , -0.10737023, -0.099812 , -0.02796177, -0.10710261,
-0.07656742, -0.07045404, -0.06649192, -0.06076612, -0.0561549 ,
-0.05177564]
compared to the original wB97X-V paper, this minimum is roughly twice as low as the original one. Do you please have got suggestions what am I doing wrong?
using Psi4 v 1.3rc2 installed with anaconda.
Thank you.
Kind regards,
stanislav.